Twice over the past several decades, researchers at Iceland's deCODE Genetics have generated maps of the human genome that have helped geneticists to study our biological blueprints. The latest version, Icelandic genome 3.0, one might call it, was published recently in Science and is the most detailed map to date. It has allowed the researchers to gain additional insight into how evolution occurs and how new diseases emerge.
"The classic premise of evolution is that it is powered first by random genetic change. But we see here in great detail how this process is in fact systematically regulated – by the genome itself and by the fact that recombination and de novo mutation are linked," said senior author Kári Stefánsson, MD, CEO of deCODE, a division of Amgen, and the University of Iceland in Reykjavik. (see short interview with Stefánsson and press release)
First author Bjarni V. Halldórsson, also of deCODE and the University of Reykjavik, and colleagues had access to genomic data from about 150,000 Icelanders, spanning several generations, allowing them to identify new recombinations--the erroneous exchange of genomic material between strands of DNA when sperm and eggs divide--as well new mutations. Far from occurring randomly throughout the genome, it turns out that new mutations are 50 times more likely to occur in the neighborhood of recombination hotspots.
Both recombinations and mutations contribute to the diversity of humankind, but the flip side of the coin is that new mutations cause new diseases, especially the debilitating and often fatal rare diseases of early childhood. Stefánsson's group found that women and men contribute differently to these engines of diversity: more recombinations arise from eggs and more new mutations from men. The paper also identifies several regions of the genome that have especially high recombination rates.
Armed with this new map, genetic scientists will be able to develop new leads to combat human disease.
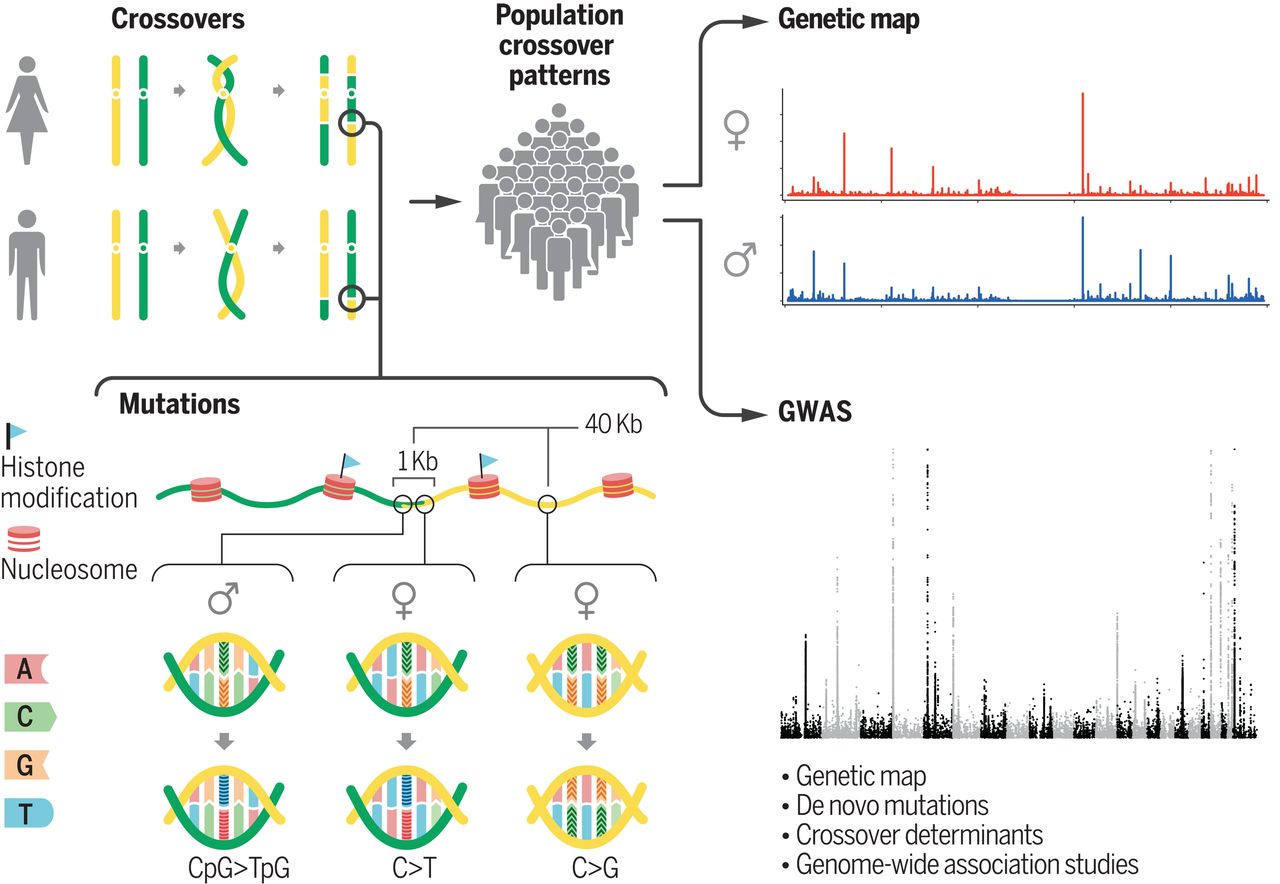
Our search for crossovers in parents and their offspring. Histone modifications influence crossover location. The DNM rate is higher within 1 kb from a crossover in both sexes, but the type of mutations differs between the sexes. The DNM rate is also higher up to 40 kb from crossovers in females with enrichment of G→C mutations. We used crossovers from many individuals to construct genetic maps and performed genome-wide association studies (GWAS) on the recombination rate and attributes of crossover locations to search for genes that control crossover characteristics. (From Halldorsson et al., 2019.)
Reference
Halldorsson BV, Palsson G, Stefansson OA, Jonsson H, Hardarson MT, Eggertsson HP, Gunnarsson B, Oddsson A, Halldorsson GH, Zink F, Gudjonsson SA, Frigge ML, Thorleifsson G, Sigurdsson A, Stacey SN, Sulem P, Masson G, Helgason A, Gudbjartsson DF, Thorsteinsdottir U, Stefansson K. Characterizing mutagenic effects of recombination through a sequence-level genetic map. Science. 2019 Jan 25;363(6425). Erratum in: Science. 2019 Feb 8;363(6427).

