By Carol Cruzan Morton
29 November 2020. Sorting out the genetics of disease risk has been challenging, but it may turn out to be the easier part of developing medically useful products and information in the upcoming era of precision medicine, according to Kári Stefánsson, founder and CEO of deCODE genetics in Iceland.
Stefánsson made his remarks in the first of the Nordic Society of Human Genetics and Precision Medicine (NSHG-PM) 2020-2021 webinar series, on 9 November 2020. He gave a pre-recorded talk on deCODE’s multi-omics approach, followed by a live question-and-answer session with an expert panel.
Genetic association studies use two data sets, one with diverse genome sequences and the other with diverse phenotypes. To follow the genomic clues to the underlying biology and pathogenesis, deCODE has added more data sets to the analytical mix: RNA transcripts and proteins, as measured in blood plasma.
“It's extremely complicated, particularly when you look at variants in the genome that affect the levels of many proteins,” Stefánsson said. “When it comes to analyzing the genome, you can basically use the same methods on all variants. When it comes to the proteome, you cannot use the same approach in the analysis of every protein, or in the change in the levels of all proteins. It becomes complicated, but very exciting, very interesting.”
Last year, Stefánsson was chosen the first president of the Society, which formed to establish a framework for human genetics research and the application of genomics to healthcare across the region, with the aim of generating and integrating genomic and healthcare data from Iceland, Denmark, Estonia, Finland, Norway, and Sweden.
Hypothesis-free research
The talk covered the population-scale genetics approach Stefánsson pioneered at deCODE to understand the wide spectrum of diversity in the human genome and to explore how diseases come about and how they may respond to treatments. Discussion topics ranged from science to Nordic strategies to lead the way in precision medicine.
The ability to scan the entire genome has led to an “avalanche of discoveries,” in part by opening up a new hypothesis-independent approach, he said.
“Once you get into a position where you can begin to explore the data without a clear idea of what the results may be, it opens up all kinds of opportunities,” he said in answer to a question. “I think that one of the important aspects of a well-designed population approach is to gather your data independent of the questions that you might ask from them. There is a temptation to let the questions dictate how you gather the data.”
The transformative nature of hypothesis-independent research is illustrated by atrial fibrillation, a common cause of stroke and heart failure. All of the risky genetic variants deCODE found have to do with the structure of the heart, suggesting the associated arrhythmia may be secondary to atrial cardiomyopathy, instead of originating with ion channels and the heart’s electrical mechanism, Stefánsson said.
There are implications for treatment already in these findings, and evidence of the value of this sort of hypothesis-free population level research, he noted. But it also sets the stage for bring other -omics techniques to bear.
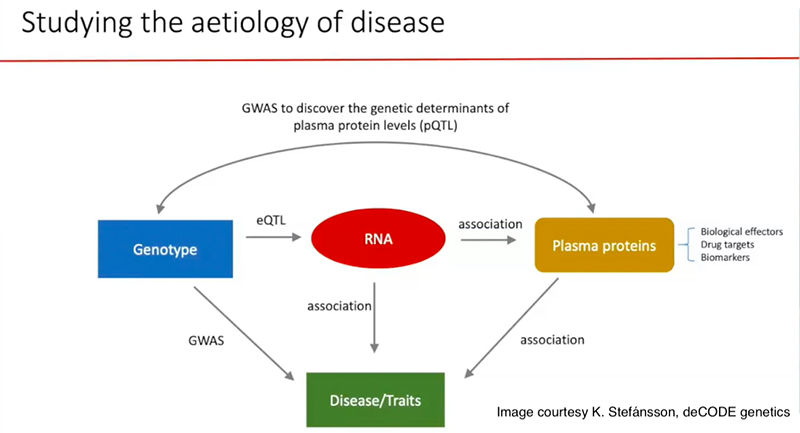
Omics at different levels
 Describing work in progress on the APOE gene and risk of Alzheimer’s disease, Stefánsson showed the power of adding just one other -omics approach—plasma protein screens—to the genetics. It appears that the levels of nearly 300 proteins are regulated by variants in of the APOE protein, raising that the possibility that the altered AD risk does not arise directly via the well-known e2 or e4 variants.
Describing work in progress on the APOE gene and risk of Alzheimer’s disease, Stefánsson showed the power of adding just one other -omics approach—plasma protein screens—to the genetics. It appears that the levels of nearly 300 proteins are regulated by variants in of the APOE protein, raising that the possibility that the altered AD risk does not arise directly via the well-known e2 or e4 variants.
“Our data indicate that the sequence variants in APOE influence the risk of Alzheimer’s disease through totally different proteins,” Stefánsson said.
For its proteomic analyses, deCODE deploys a SomaLogic assay that can assess 5,000 proteins at once. In general, Stefánsson said, they are finding that most proteins in plasma are influenced by at least one sequence variant, and some by many variants. On the flip side, most gene variants influence the level of only one protein, while others can change the levels of more than 100 proteins.
“These generate a complexity when you're trying to figure out which one of the proteins is responsible for a component of the phenotype that you're interested in,” he said. “It is very important to recognize that once you find an association between the level of a protein and the risk of a disease, you have to realize that there are two possibilities. It is more often that you see the plasma level of proteins changed as a consequence of the disease, rather than the disease being a consequence of changes in the level of the protein.”
The most complete example of how deCODE’s multi-omics approach can illuminate pathogenic mechanisms was its recent paper on autoimmune thyroiditis, said webinar panelist Mark Daly, director of the Institute for Molecular Medicine Finland (FIMM).
Using 30,000 cases and 700,000 controls, the deCODE team found 99 risk variants (see NSHG-PM news https://www.nshg-pm.org/Iceland-Omics-IDs-Autoimmune-Risk-Gene). The largest risk came from the FLT3 gene. RNA data showed the variant produced a shorter spliced RNA and ultimately a truncated protein with a loss of a key function. But in a puzzle, the same genetic change was known to be an activating somatic mutation in patients with acute myelogenous leukemia. A proteome analysis resolved the apparent contradiction, showing double the levels of FLT3 ligand in carriers of the variant, resulting in an overall gain of function.
In other work with this new proteomics approach, deCODE has calculated that a “relatively small” group of proteins has the power to predict the remaining lifespan among people between ages 60 and 80.
At one extreme, these proteins identify a group of 5% of people who have about 90% probability of dying within the next five years, and at the other extreme, another 5% that has virtually no probability of dying within the next five years, Stefánsson said.
The protein assay also correlates well with measures of human frailty such as hand grip, exercise tolerance, and cognitive function, he added.
Beyond the effects of common variation, Stefánsson stressed the need to study de novo mutations and their effects on risk of disease. In particular, he described the growing support for hypotheses that tie increased paternal age to common diseases as varied as schizophrenia and cancer. This contrasts with the more well-known, but more rare, maternal risk from large chromosomal rearrangements.
Progress in understanding these events, as well as the gradual changes due to selection, will have to come from more basic research about human genetics and even evolutionary genetics.
Panel stresses Nordic opportunities
Panelists Paul Franks of Lund University, Søren Brunak of the University of Copenhagen, and Ole Andreassen of the University of Oslo also asked Stefánsson for a perspective on the next steps and priorities for moving from human genetics to precision medicine.
“The most significant bottleneck for all of us, when it comes to discovering something about the nature of human disease today, is access to material,” Stefánsson said. “We have to figure out a way in which we can get access to clinical material in such a way that society in general feels at ease with it.”
The Nordic countries have unique advantages, such as a single-payer health system, health care registries, and a population with a tradition of participating in biomedical research. Around the world, he said, when people recover from an illness or find relief from worrisome symptoms, they usually thank hospital staff and health professionals. But they can also credit decades of Nordic epidemiological data used to make a discovery that was turned into a way to treat or prevent a disease.
That data is even more important in delivering on the promise of personalized medicine, which promises the potential of more health benefits for individuals and presents more complex research challenges, Stefánsson said. “In the Nordic countries, we have an opportunity to lead the world in this kind of biomedical research.”
The webinar attendance was about 115 people, said Hakon Heimer of the University of Copenhagen and executive officer of the society. The society plans regular webinars over the winter for its members, which includes researchers, clinicians, data scientists, and others engaged in building a research base for precision medicine and translating it into clinical practice.

